
Protein Identification and Quantification
DIA-NN
DIA - Data are even larger than DDA data. The database search requires more computing power. Such a computational task should be limited to larger computers. Here, we present only the DIA-NN interface and how the search is done.
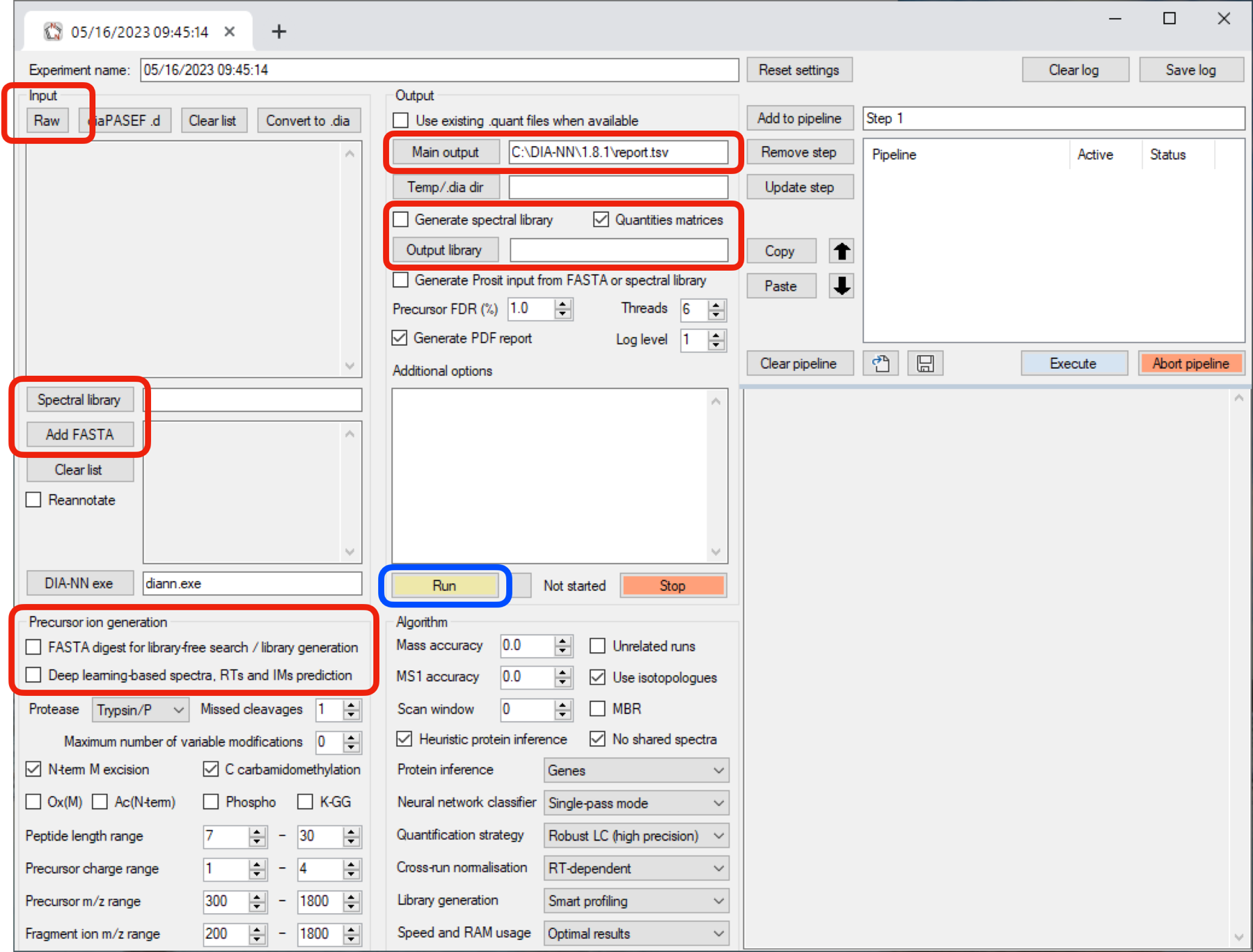
- You declare the raw data files by clicking the Raw button (for .wiff and .raw formats additional software is required - see Installation)
- You add a FASTA protein sequence database by using the Add FASTA button.
- The DIA data are matched against a spectral library, which you can declare by hitting the Spectral Library button - or the spectral library is calculated internally from the FASTA protein sequence database by activating the FASTA digest for library-free search / library generation option.
- You set the main output folder (away from the currently set installation folder) using the Main output button
- Optionally you generate a spectral library by activating the Generate spectral library option and by defining its location using the Output library button.
- The search is startet by hitting the Run button.
Results
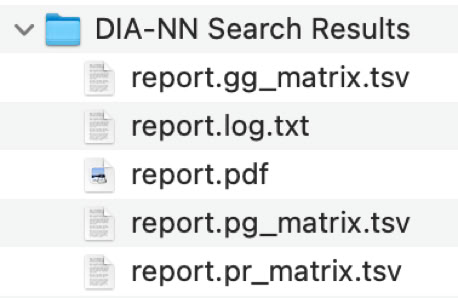
See the Manual page for a detailed discussion of the content of the different result files. Here, the protein groups file "report.pg_matrix.tsv" will be used for further analysis.
Comments: matthias.wilm@ucd.ie

