

Parameters to set:
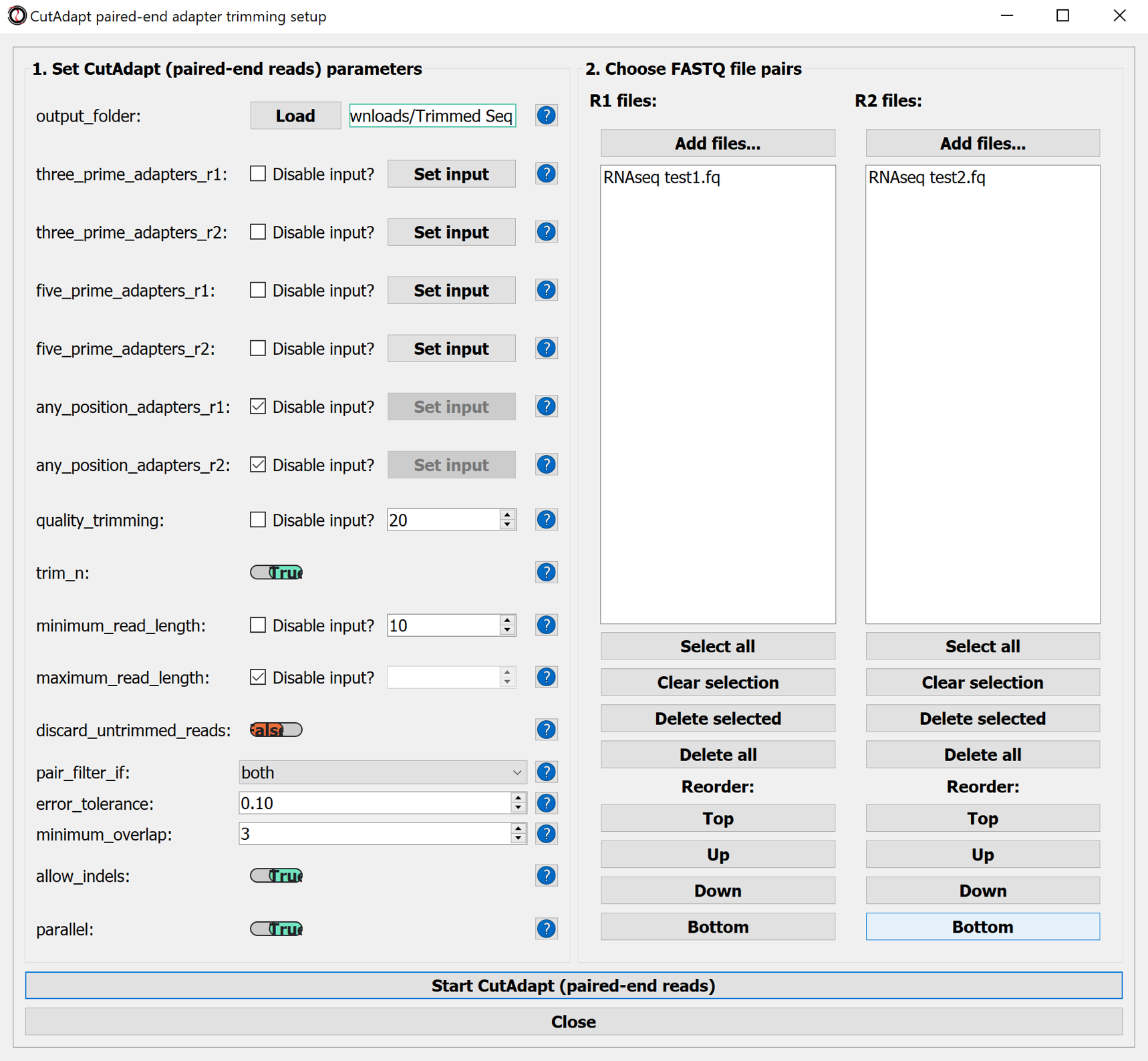
- output_folder
- R1 files and matching R2 files : the RNAseq data files
- Adapter sequences:
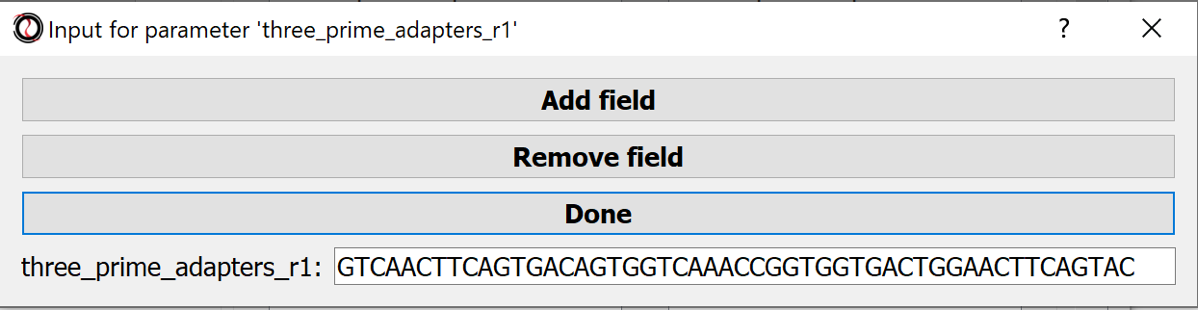
- three_prime_adapters_r1, - three_prime_adapters_r2, optionally: other adapter sequences
- Follow the original manual for the other parameters
- Use the button Start CutAdapt (paired-end reads) to start trimming
- Use the "Resource Monitor" application to monitor computer activity
Results:
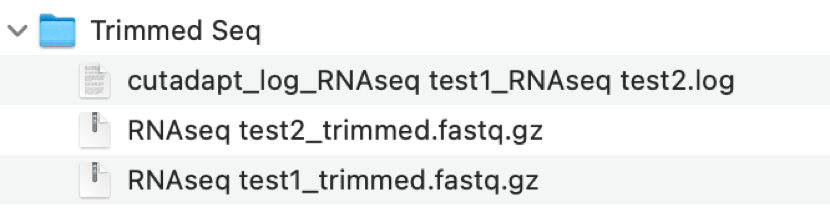
The result files should have roughly the same size as the untrimmed data files. Trimmed data files can be used to quantify the RNA expression levels.
Comments: matthias.wilm@ucd.ie
.