
The tables of over- and under-expressed genes can be used for pathway analysis. Save them, open them in a spreadsheet programme, copy the desired gene names from the gene names column and use them in pathway analysis programmes. One possibility is the string database: https://string-db.org




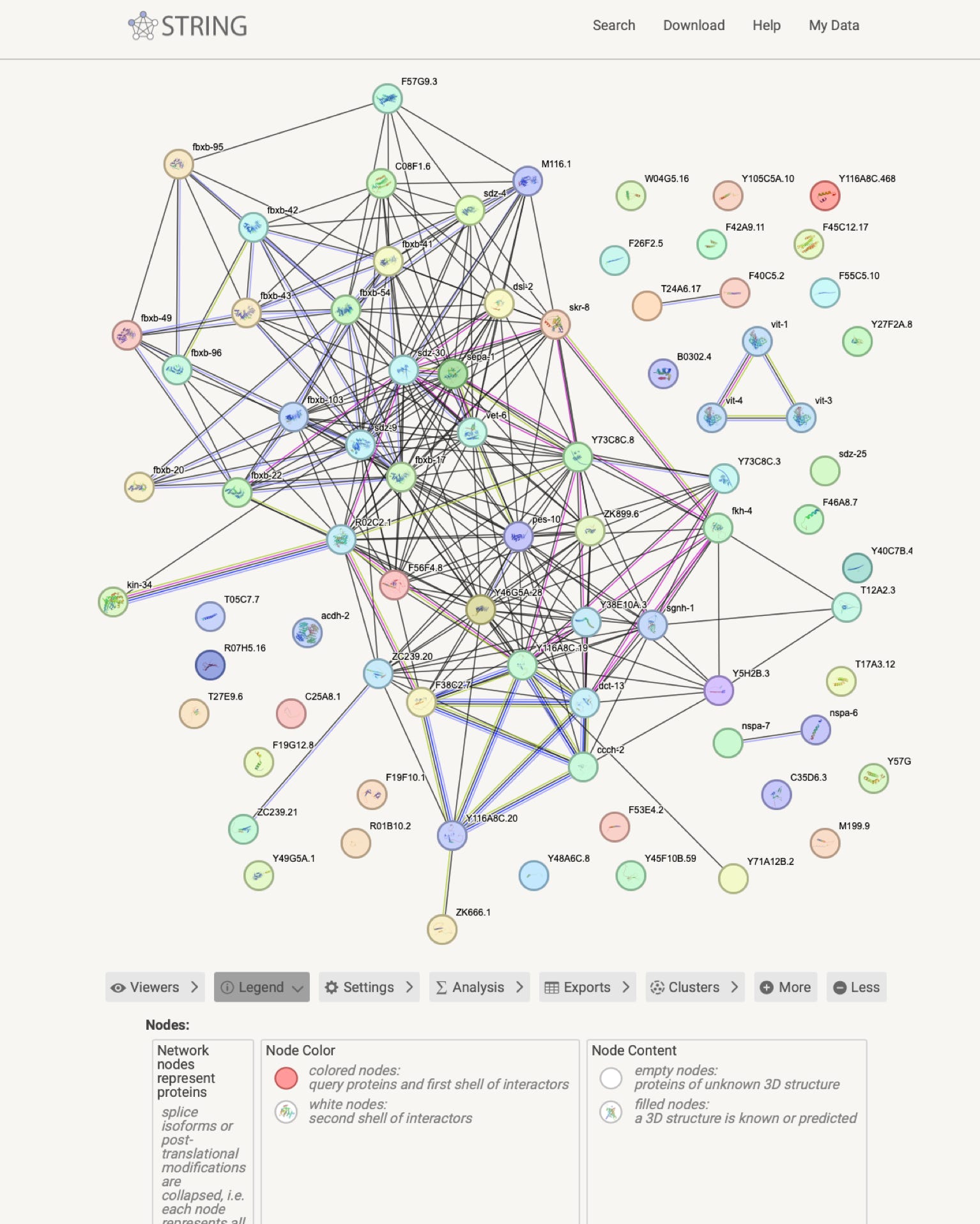
The string website allows then extensive analysis of the pathways retrieved.
Where to go from here ?
The differential analysis scratches just the surface of what RNAlysis can do for you.
It can handle much more complex experimental situations (see the original tutorial or the complete manual).
Examples are:
- Grouping over- and under-expressed genes in several experimental conditions in form of a Venn diagram.
- Checking whether genes with a particular attribute are preferable over- oder under-expressed (enrichment analysis).
- Time series analysis.
- Cluster analysis etc.
If you use it, don't forget to reference the contributors to this software.
Comments: matthias.wilm@ucd.ie
.