

- R: A statistical analysis package: Source - Installer (ARM chip) - Installer (Intel chip)
- R Package: DeSeq2 (needs to be installed from the running the R-programme in macOS): Source -
Installation: Run the following code from the R-command line after having started the R programme from the
Applications folder:
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("DESeq2")
- Kallisto : Python script to quantify RNAseq data: Source - Installer: copy the files to the "/Applications/" folder
- RNAlysis : RNAseq data analysis programme: Source - Installer: copy the decompressed RNAlysis file to the
"/Applications/" folder
- FastQC : Sequence quality check programme: Source - Installer: Double click the dmg file and move the FastQC application to the "/Applications/" folder
- Java Runtime Environment: Required to run FastQC: Source - Installer (ARM chip) - Installer (Intel chip) : Double-click the installer and follow instructions
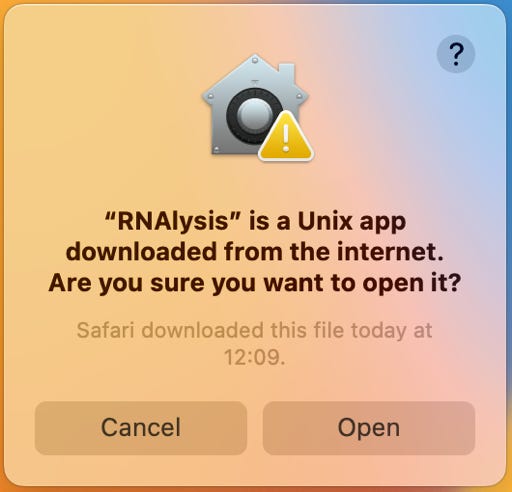
First run of RNAlysis and of FastQC: RNAlysis and FastQC are not signed by a registered Apple Developer. You have to allow the system to run the application:
- "control-click" RNAlysis or FastQC and choose the context menu: Open
- click the Open button
- wait a bit
Comments: matthias.wilm@ucd.ie