

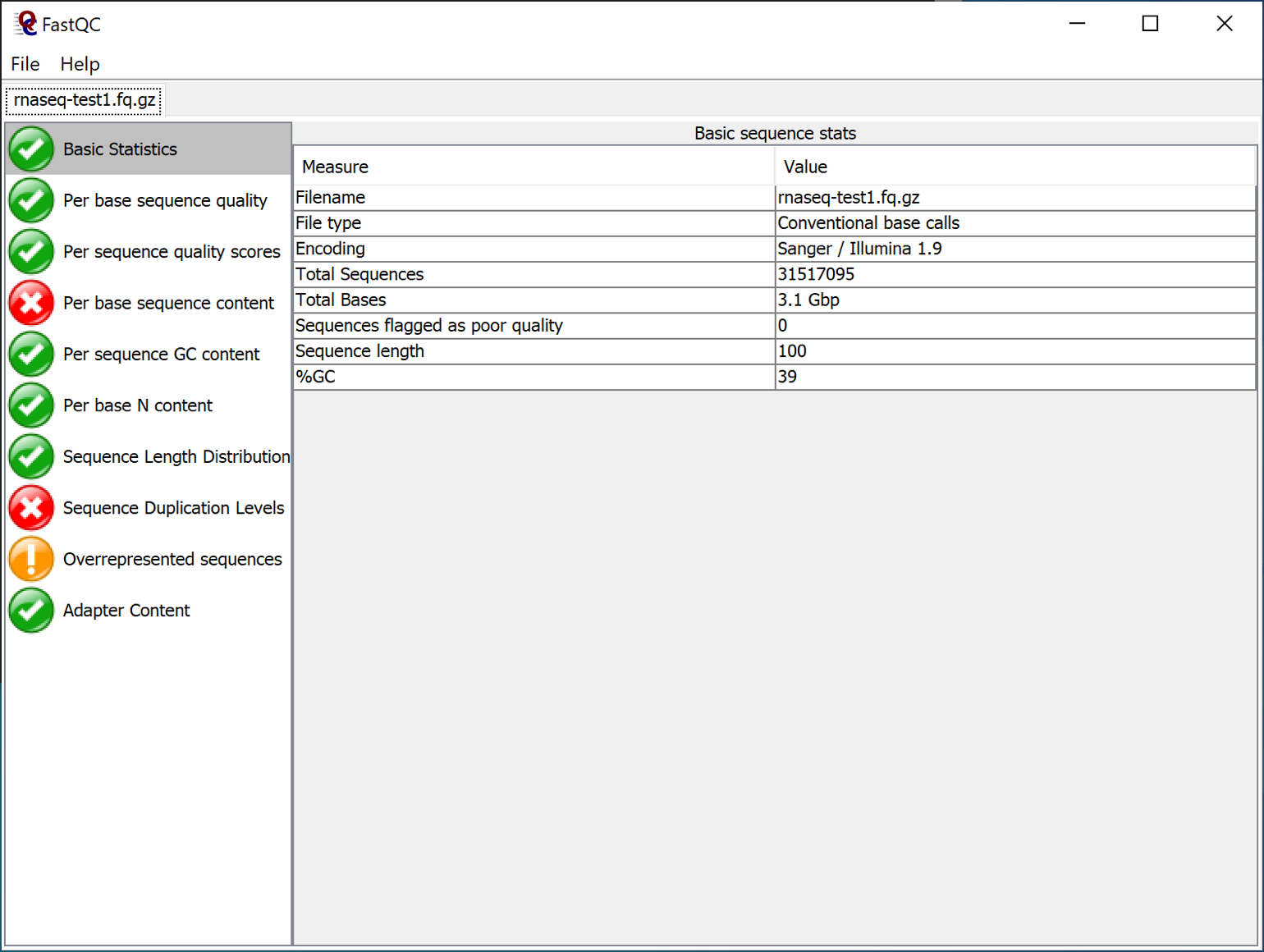
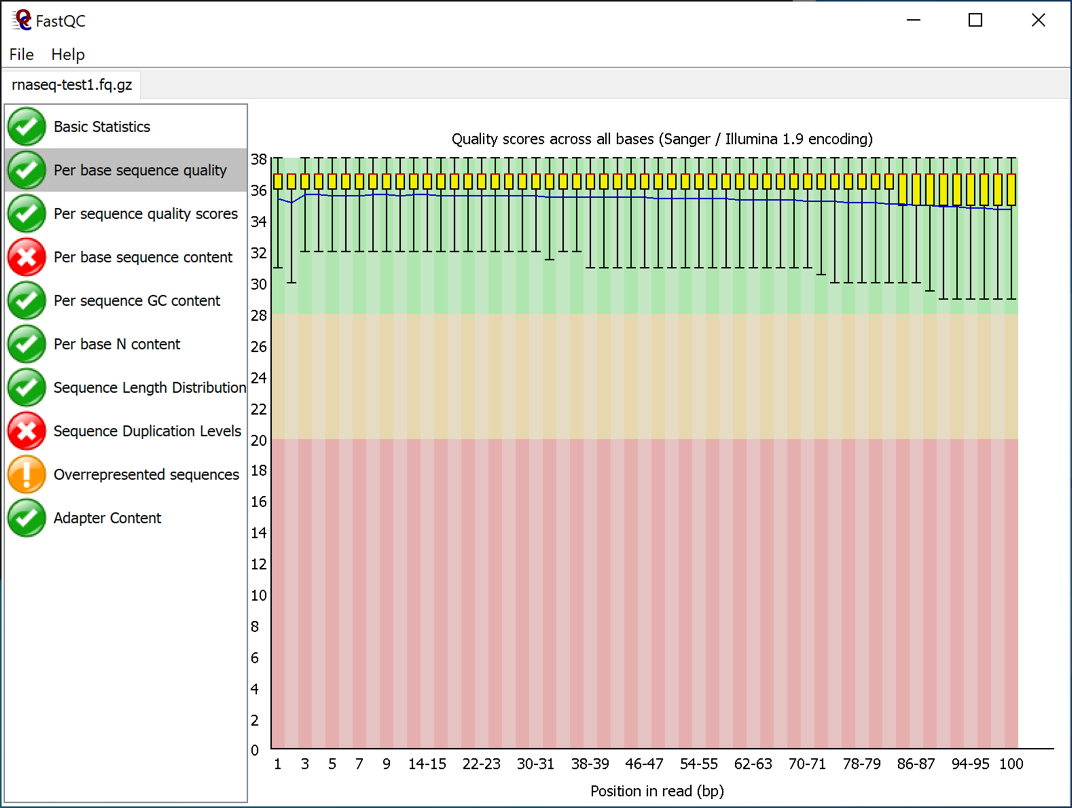
Open a sequence file (for instance the "rnaseq-test1.fq.gz" file).
Command: File > Open...
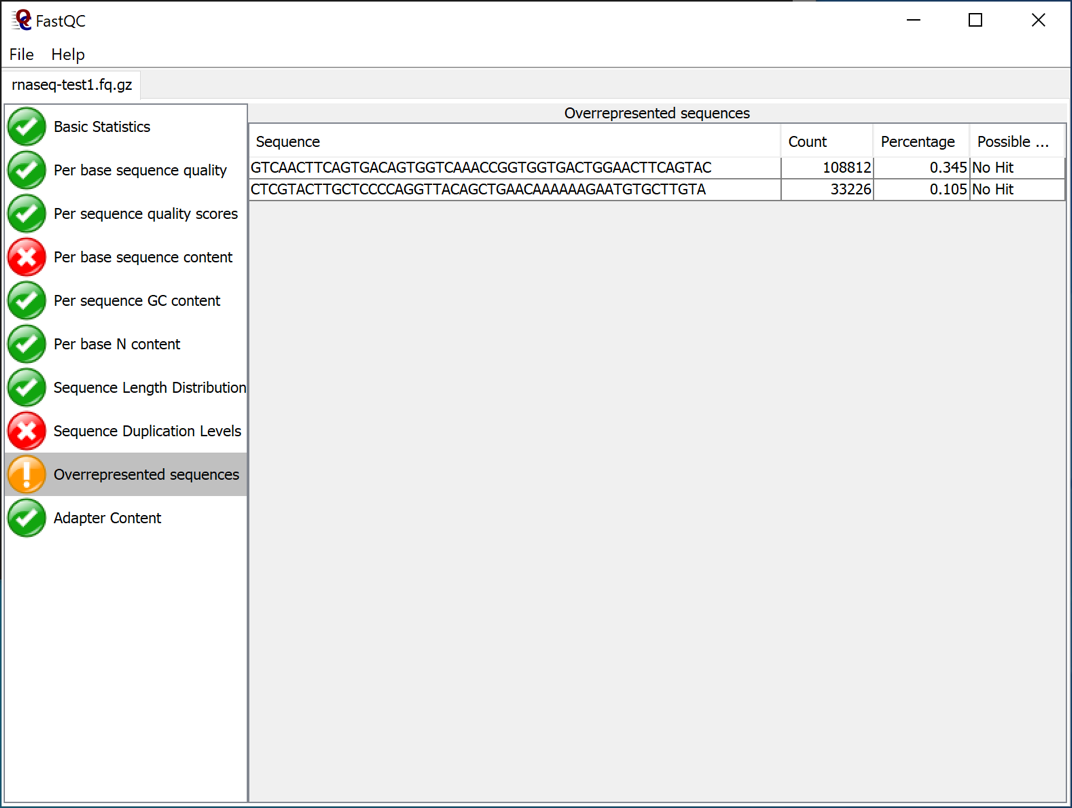
Results: The "Overexpressed Sequences" pane shows sequences that are likely to be adapter sequences from the sequencing reactions.
Comments: matthias.wilm@ucd.ie
.